STCRpy
Software suite for T-cell receptor structures
Software suite for T-cell receptor structures
ImmuneBuilder for T-cell receptor structures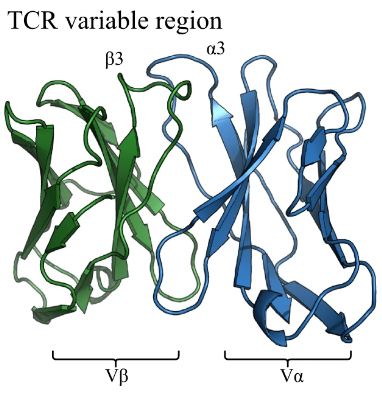
Implementation for calculating root-mean-square deviation of length-mismatched proteins via spline upsampleing and dynamic time warping.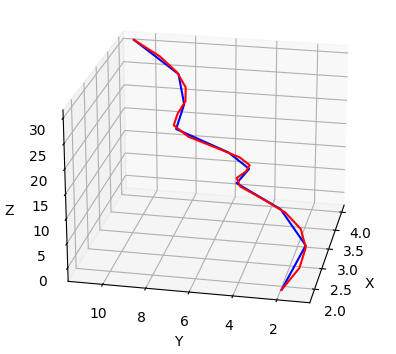
Prototype Application for privacy-preserving contact tracing in pandemic scenarios.
This is an adapted beamer template for generating academic slides.
Published in ArXiv, 2020
Recommended citation: Holzapfel et. al. (2024). "Digital Contact Tracing Service: An improved decentralized design for privacy and effectiveness." ArXiv. 1(1).
Download Paper
Published in Cell Reports, 2024
Recommended citation: Raybould & Greenshields-Watson et. al. (2024). "The Observed T cell receptor Space database enables paired-chain repertoire mining, coherence analysis and language modeling." Cell Reports. 43(9).
Download Paper
Published in Machine Learning for Structural Biology @ NeurIPS 2024, 2024
Recommended citation: Quast & Sweeney et. al., (2024). "Estimating protein flexibility via uncertainty quantification of structure prediction models." Machine Learning for Structural Biology @ NeurIPS 2024. 1(1).
Download Paper
Published in Communications Biology, 2025
Recommended citation: Quast et. al., (2009). "T-cell receptor structures and predictive models reveal comparable alpha and beta chain structural diversity despite differing genetic complexity." Communications Biology. 8(1).
Download Paper
Published in BioRxiv, 2025
Recommended citation: Quast et.al. (2025). "STCRpy: a software suite for T cell receptor structure parsing, interaction profiling and machine learning dataset preparation." BioRxiv. 1(1).
Download Paper
Published:
Presented a poster: “A Structure-Focused Analysis of Antigens Presented by the Class 1 Major Histocompatibility Complex”, which won the 3D-Sig RSCB PDB Award for best poster, at ISMB 2023.
Published:
Presented a poster “T-cell receptor structures and predictive models reveal comparable alpha and beta chain structural diversity despite differing genetic complexity” at the conference: Adaptive Immune Receptors: Structural Modelling and Immunoinformatics
Published:
I presented a suite of machine learning tools for in-silico Antibody design and developability as part of the “In silico and Machine Learning Tools for Antibody Design and Developability Predictions” series.
Published:
Presented a poster on joint first author workshop paper: “Estimating protein flexibility via uncertainty quantification of structure prediction models” at the Machine Learning for Structural Biology Workshop at NeurIPS 2024.
MSc and BSc Thesis Supervision, University of Oxford & University of Stuttgart, 2025
======
Lectures, workshops and tutorials, University of Oxford & University of Stuttgart, 2025
======