Sitemap
A list of all the posts and pages found on the site. For you robots out there, there is an XML version available for digesting as well.
Pages
Posts
OPIG Blog Posts
Published:
Currently I write blog posts on the Oxford Protein Informatics Group blog: blopig
- Cross referencing across LaTeX documents in one project
- Making your code pip installableMaking your code pip installable
- Taking Equivariance in deep learning for a spin?
- How to replace bike ball bearings when your steering sounds crunchy
- LaTeX Beamer Template with Logos
- Graphormer: Merging GNNs and Transformers for Cheminformatics
portfolio
STCRpy
Software suite for T-cell receptor structures
TCRBuilder2+
ImmuneBuilder for T-cell receptor structures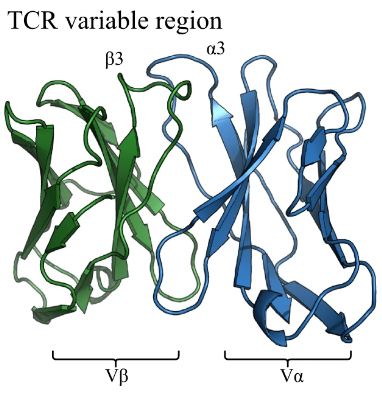
RMSD for length-mismatched protein structures
Implementation for calculating root-mean-square deviation of length-mismatched proteins via spline upsampleing and dynamic time warping.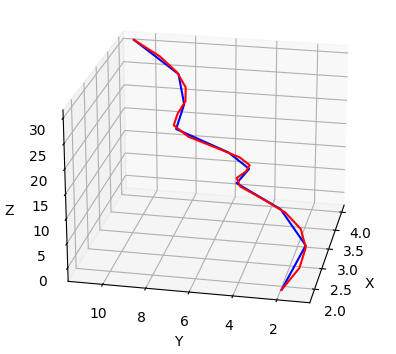
ito app
Prototype Application for privacy-preserving contact tracing in pandemic scenarios.
Academic slides template in latex
This is an adapted beamer template for generating academic slides.
publications
Digital contact tracing service: An improved decentralized design for privacy and effectiveness
Published in ArXiv, 2020
Recommended citation: Holzapfel et. al. (2024). "Digital Contact Tracing Service: An improved decentralized design for privacy and effectiveness." ArXiv. 1(1).
Download Paper
The Observed T cell receptor Space database enables paired-chain repertoire mining, coherence analysis and language modeling
Published in Cell Reports, 2024
Recommended citation: Raybould & Greenshields-Watson et. al. (2024). "The Observed T cell receptor Space database enables paired-chain repertoire mining, coherence analysis and language modeling." Cell Reports. 43(9).
Download Paper
Estimating protein flexibility via uncertainty quantification of structure prediction models
Published in Machine Learning for Structural Biology @ NeurIPS 2024, 2024
Recommended citation: Quast & Sweeney et. al., (2024). "Estimating protein flexibility via uncertainty quantification of structure prediction models." Machine Learning for Structural Biology @ NeurIPS 2024. 1(1).
Download Paper
T-cell receptor structures and predictive models reveal comparable alpha and beta chain structural diversity despite differing genetic complexity
Published in Communications Biology, 2025
Recommended citation: Quast et. al., (2009). "T-cell receptor structures and predictive models reveal comparable alpha and beta chain structural diversity despite differing genetic complexity." Communications Biology. 8(1).
Download Paper
STCRpy: a software suite for T cell receptor structure parsing, interaction profiling and machine learning dataset preparation
Published in BioRxiv, 2025
Recommended citation: Quast et.al. (2025). "STCRpy: a software suite for T cell receptor structure parsing, interaction profiling and machine learning dataset preparation." BioRxiv. 1(1).
Download Paper
talks
A Structure-Focused Analysis of Antigens Presented by the Class 1 Major Histocompatibility Complex
Published:
Presented a poster: “A Structure-Focused Analysis of Antigens Presented by the Class 1 Major Histocompatibility Complex”, which won the 3D-Sig RSCB PDB Award for best poster, at ISMB 2023.
T-cell receptor structures and predictive models reveal comparable alpha and beta chain structural diversity despite differing genetic complexity
Published:
Presented a poster “T-cell receptor structures and predictive models reveal comparable alpha and beta chain structural diversity despite differing genetic complexity” at the conference: Adaptive Immune Receptors: Structural Modelling and Immunoinformatics
PEGS 2024
Published:
I presented a suite of machine learning tools for in-silico Antibody design and developability as part of the “In silico and Machine Learning Tools for Antibody Design and Developability Predictions” series.
Estimating protein flexibility via uncertainty quantification of structure prediction models
Published:
Presented a poster on joint first author workshop paper: “Estimating protein flexibility via uncertainty quantification of structure prediction models” at the Machine Learning for Structural Biology Workshop at NeurIPS 2024.
teaching
Supervision
MSc and BSc Thesis Supervision, University of Oxford & University of Stuttgart, 2025
======
Teaching
Lectures, workshops and tutorials, University of Oxford & University of Stuttgart, 2025
======